
To understand if cDNA-starts can be used to assign RNA-binding sites, we further analysed the iCLIP data with high frequency of non-coinciding cDNA-starts. The study concluded that the use of cDNA-starts may not be appropriate in iCLIP whenever non-coinciding cDNA-starts are prevalent. The non-coinciding cDNA-starts in eIF4A3 iCLIP data produced by the previous study were shifted upstream of this expected region and it was proposed that the presence of non-coinciding cDNA-starts might be related to this shift. However, further studies showed that the sequence and structure of a nascent messenger RNA (mRNA) can shift EJC deposition as far as 10 nt away from this expected site. In vitro biochemical experiments with several splicing substrates demonstrated that the site of EJC deposition is normally expected at nucleotides −20 to −24 upstream of the exon-exon junction (–24.–20 nt). eIF4A3 is a component of the exon junction complex (EJC). Here, we focused on experiments produced for polypyrimidine tract binding protein 1 (PTBP1), eukaryotic initiation factor 4A-III (eIF4A3) and the splicing factor U2 auxiliary factor 65 kDa subunit (U2AF2), which represent examples of non-coinciding or coinciding cDNA-starts in introns or exons. However, a recent study observed that the starts of long and short iCLIP cDNAs often map to different genomic positions for several RBPs, which leads to non-coinciding cDNA-starts. The computational methods that use cDNA-starts to assign RNA-binding sites have been developed along with iCLIP. Therefore, understanding the proportion and characteristics of truncated cDNAs in these protocols is essential. Recently, further variants were developed that also amplify truncated cDNAs, including BrdU-CLIP, eCLIP and irCLIP. Even though iCLIP amplifies both truncated and readthrough cDNAs, computational comparisons of CLIP and iCLIP cDNAs estimated that over 80% of iCLIP cDNAs truncate at the crosslink sites of most RBPs. The cDNA-starts of these truncated cDNAs identify the nucleotide just downstream of the crosslinked peptide. Therefore, individual-nucleotide resolution CLIP (iCLIP) was also developed to exploit the ‘truncated cDNAs’. The CLIP protocol prepares the cDNA library in a way that requires the reverse transcriptase to read through this peptide, thereby generating only ‘readthrough cDNAs’. This is followed by reverse transcription, during which the bound peptide can lead to truncation of complementary DNAs (cDNA) at the crosslink site. During the CLIP protocol, crosslinked protein–RNA complexes are purified and the RNA fragments are released by digesting the protein, resulting in RNAs with a covalently bound peptide at the crosslink site. To understand the mechanisms of their action, it is essential to identify the endogenous sites of protein–RNA interactions, which has been aided by the development of ultraviolet (UV) crosslinking and immunoprecipitation (CLIP). RNA-binding proteins (RBPs) play crucial roles in all aspects of post-transcriptional gene regulation. We demonstrate the advantage of iCLIP and related methods that can amplify cDNAs that truncate at crosslink sites and we show that computational analyses based on cDNAs-starts are appropriate for such methods. In contrast, we show that a broad size range of cDNAs in iCLIP allows the cDNA-starts to efficiently delineate the complete RNA-binding sites. Our study also shows that if RNase does not efficiently cut within the binding sites, the original CLIP method is less capable of identifying the longer binding sites of RBPs. These constraints are overcome when fragmentation by RNase I is efficient and when a broad cDNA size range is obtained. As previously noted, the positions of complementary DNA (cDNA)-starts depend on cDNA length in several iCLIP experiments and we now find that this is caused by constrained cDNA-ends, which can result from the sequence and structure constraints of RNA fragmentation.
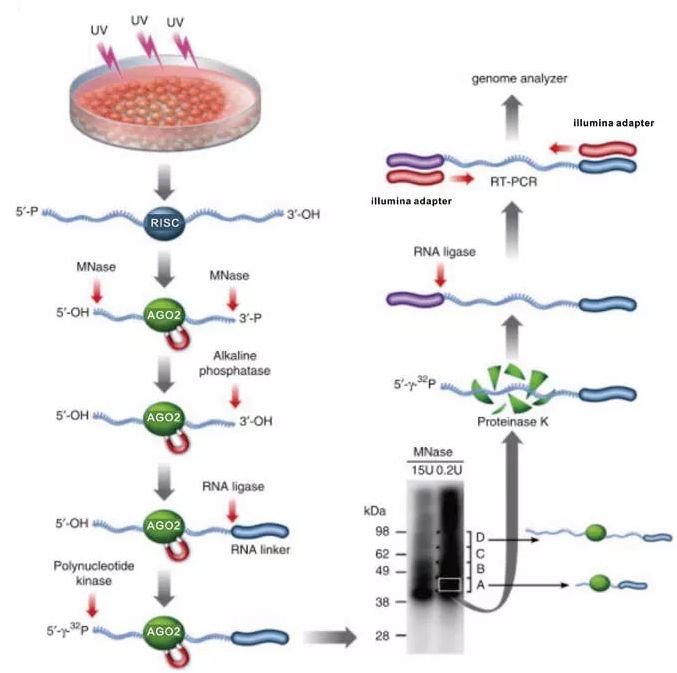
We perform experiments for PTBP1 and eIF4A3 using individual-nucleotide resolution CLIP (iCLIP), employing either UV-C or photoactivatable 4-thiouridine (4SU) combined with UV-A crosslinking and compare the results with published data. Here, we produce data with multiple variants of CLIP and evaluate the data with various computational methods to better understand their suitability. This variety of approaches can create challenges for a novice user and can hamper insights from multi-study comparisons. Several variants of CLIP exist, which require different computational approaches for analysis. Ultraviolet (UV) crosslinking and immunoprecipitation (CLIP) identifies the sites on RNAs that are in direct contact with RNA-binding proteins (RBPs).


 0 kommentar(er)
0 kommentar(er)
